What is Pandas?¶
pandas= Python Data Analysis Library- Library for working with data tables (called DataFrames in
pandas)
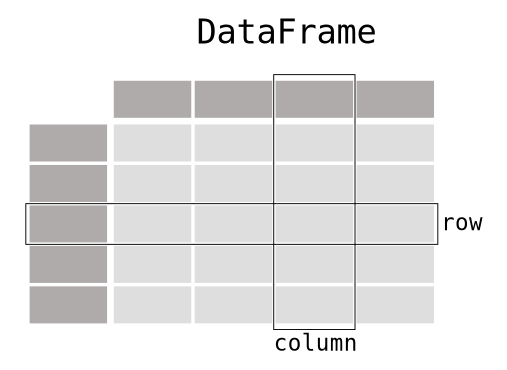
In this lesson, we'll be covering the following topics:
Additional resources:
pandas documentationpandas = Python Data Analysis Librarypandas)
pandas you can do pretty much everything you would in a spreadsheet, plus a whole lot more!First, we need to import the pandas library:
import pandas
We'll be working with data about countries around the world, from the Gapminder foundation. You can view the data table online here.
| Column | Description |
|---|---|
| country | Country name |
| population | Population in the country |
| region | Continent the country belongs to |
| sub_region | Sub regions as defined by |
| income_group | Income group as specified by the world bank |
| life_expectancy | The average number of years a newborn child would live if mortality patterns were to stay the same |
| gdp_per_capita | GDP per capita (in USD) adjusted for differences in purchasing power |
| children_per_woman | Number of children born to each woman |
| child_mortality | Deaths of children under 5 years of age per 1000 live births |
| pop_density | Average number of people per km$^2$ |
| years_in_school_men | Average number of years attending primary, secondary, and tertiary school for 25-36 years old men |
| years_in_school_women | Average number of years attending primary, secondary, and tertiary school for 25-36 years old women |
We'll use the function read_csv() to load the data into our notebook
read_csv() function can read data from a locally saved file or from a URLworldworld = pandas.read_csv('https://raw.githubusercontent.com/jenfly/datajam-python/master/data/gapminder.csv')
world
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | Asia | Southern Asia | Low | 32.0 | 1040 | 7.57 | 425.0 | 11.9 | NaN | NaN |
| 1 | Afghanistan | 1955 | 8270000 | Asia | Southern Asia | Low | 35.1 | 1130 | 7.52 | 394.0 | 12.7 | NaN | NaN |
| 2 | Afghanistan | 1960 | 9000000 | Asia | Southern Asia | Low | 38.6 | 1210 | 7.45 | 364.0 | 13.8 | NaN | NaN |
| 3 | Afghanistan | 1965 | 9940000 | Asia | Southern Asia | Low | 42.2 | 1190 | 7.45 | 334.0 | 15.2 | NaN | NaN |
| 4 | Afghanistan | 1970 | 11100000 | Asia | Southern Asia | Low | 45.8 | 1180 | 7.45 | 306.0 | 17.0 | 1.36 | 0.21 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2487 | Zimbabwe | 1995 | 11300000 | Africa | Sub-Saharan Africa | Low | 53.7 | 2480 | 4.43 | 90.1 | 29.3 | 8.41 | 6.92 |
| 2488 | Zimbabwe | 2000 | 12200000 | Africa | Sub-Saharan Africa | Low | 46.7 | 2570 | 4.06 | 96.8 | 31.6 | 9.07 | 7.71 |
| 2489 | Zimbabwe | 2005 | 12900000 | Africa | Sub-Saharan Africa | Low | 45.3 | 1650 | 3.99 | 99.7 | 33.4 | 9.73 | 8.53 |
| 2490 | Zimbabwe | 2010 | 14100000 | Africa | Sub-Saharan Africa | Low | 49.6 | 1460 | 4.03 | 89.9 | 36.4 | 10.40 | 9.36 |
| 2491 | Zimbabwe | 2015 | 15800000 | Africa | Sub-Saharan Africa | Low | 58.3 | 1890 | 3.84 | 59.9 | 40.8 | 11.10 | 10.20 |
2492 rows × 13 columns
world variableNaN values—these represent missing data (NaN = "not a number")What type of object is world?
type(world)
pandas.core.frame.DataFrame
world is a DataFrame, a data structure from the pandas libraryworld, the integer numbers in bold on the left are the DataFrame's indexpandas when loading the dataFor large DataFrames, it's often useful to display just the first few or last few rows:
world.head()
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | Asia | Southern Asia | Low | 32.0 | 1040 | 7.57 | 425.0 | 11.9 | NaN | NaN |
| 1 | Afghanistan | 1955 | 8270000 | Asia | Southern Asia | Low | 35.1 | 1130 | 7.52 | 394.0 | 12.7 | NaN | NaN |
| 2 | Afghanistan | 1960 | 9000000 | Asia | Southern Asia | Low | 38.6 | 1210 | 7.45 | 364.0 | 13.8 | NaN | NaN |
| 3 | Afghanistan | 1965 | 9940000 | Asia | Southern Asia | Low | 42.2 | 1190 | 7.45 | 334.0 | 15.2 | NaN | NaN |
| 4 | Afghanistan | 1970 | 11100000 | Asia | Southern Asia | Low | 45.8 | 1180 | 7.45 | 306.0 | 17.0 | 1.36 | 0.21 |
The head() method returns a new DataFrame consisting of the first n rows (default 5)
Pro Tips!
- To display the documentation for this method within Jupyter notebook, you can run the command
world.head?or pressShift-Tabwithin the parentheses ofworld.head()- To see other methods available for the DataFrame, type
world.followed byTabfor auto-complete options
First two rows:
world.head(2)
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | Asia | Southern Asia | Low | 32.0 | 1040 | 7.57 | 425.0 | 11.9 | NaN | NaN |
| 1 | Afghanistan | 1955 | 8270000 | Asia | Southern Asia | Low | 35.1 | 1130 | 7.52 | 394.0 | 12.7 | NaN | NaN |
What do you think the tail method does?
world.tail(3)
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2489 | Zimbabwe | 2005 | 12900000 | Africa | Sub-Saharan Africa | Low | 45.3 | 1650 | 3.99 | 99.7 | 33.4 | 9.73 | 8.53 |
| 2490 | Zimbabwe | 2010 | 14100000 | Africa | Sub-Saharan Africa | Low | 49.6 | 1460 | 4.03 | 89.9 | 36.4 | 10.40 | 9.36 |
| 2491 | Zimbabwe | 2015 | 15800000 | Africa | Sub-Saharan Africa | Low | 58.3 | 1890 | 3.84 | 59.9 | 40.8 | 11.10 | 10.20 |
pandas provides many ways to quickly and easily summarize your data:
Number of rows and columns:
world.shape
(2492, 13)
world has 2492 rows and 14 columnsworld.shapeshape is a data attribute of the variable worldGeneral information about the DataFrame can be obtained with the info() method:
world.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 2492 entries, 0 to 2491 Data columns (total 13 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 country 2492 non-null object 1 year 2492 non-null int64 2 population 2492 non-null int64 3 region 2492 non-null object 4 sub_region 2492 non-null object 5 income_group 2492 non-null object 6 life_expectancy 2492 non-null float64 7 gdp_per_capita 2492 non-null int64 8 children_per_woman 2492 non-null float64 9 child_mortality 2492 non-null float64 10 pop_density 2492 non-null float64 11 years_in_school_men 1780 non-null float64 12 years_in_school_women 1780 non-null float64 dtypes: float64(6), int64(3), object(4) memory usage: 253.2+ KB
If we just want a list of the column names, we can use the columns attribute:
world.columns
Index(['country', 'year', 'population', 'region', 'sub_region', 'income_group',
'life_expectancy', 'gdp_per_capita', 'children_per_woman',
'child_mortality', 'pop_density', 'years_in_school_men',
'years_in_school_women'],
dtype='object')
The describe() method computes simple summary statistics for a DataFrame:
world.describe()
| year | population | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|
| count | 2492.00000 | 2.492000e+03 | 2492.000000 | 2492.000000 | 2492.000000 | 2492.000000 | 2492.000000 | 1780.000000 | 1780.000000 |
| mean | 1982.50000 | 2.667661e+07 | 62.567135 | 10502.302167 | 4.353752 | 105.670024 | 118.014013 | 7.687713 | 6.959556 |
| std | 20.15969 | 1.037838e+08 | 11.518029 | 15478.942158 | 2.049655 | 98.615582 | 372.055683 | 3.242840 | 3.932678 |
| min | 1950.00000 | 2.500000e+04 | 23.800000 | 247.000000 | 1.120000 | 2.200000 | 0.502000 | 0.900000 | 0.210000 |
| 25% | 1965.00000 | 1.730000e+06 | 54.200000 | 1930.000000 | 2.360000 | 24.000000 | 14.275000 | 5.097500 | 3.600000 |
| 50% | 1982.50000 | 5.270000e+06 | 64.650000 | 4925.000000 | 4.340000 | 72.400000 | 44.850000 | 7.635000 | 6.990000 |
| 75% | 2000.00000 | 1.600000e+07 | 71.600000 | 12700.000000 | 6.300000 | 167.000000 | 108.000000 | 10.100000 | 10.000000 |
| max | 2015.00000 | 1.400000e+09 | 83.800000 | 178000.000000 | 8.870000 | 473.000000 | 7910.000000 | 15.300000 | 15.700000 |
The describe() method is a convenient way to quickly summarize the averages, extremes, and variability of each numerical data column.
You can look at each statistic individually with methods such as mean(), median(), min(), max(),std(), and count()
a) Initial setup (you can skip to part b if you've already done this):
pandas librarypandas.read_csv() to read data from 'https://raw.githubusercontent.com/jenfly/datajam-python/master/data/gapminder.csv' and store it in a DataFrame called world.b) Based on the output of world.info(), what data type is the pop_density column?
c) Based on the output of world.describe(), what are the minimum and maximum years in this data?
For b) and c) you can create a Markdown cell in your notebook and write your answer there.
We can save our data locally to a CSV file using the to_csv() method:
world.to_csv('data/gapminder_world_data.csv', index=False)
This creates a file called gapminder_world_data.csv within the data sub-folder
By default, the to_csv() method will save the DataFrame's index as an additional column in the CSV file. To turn this off, we use the keyword argument index=False.
Let's check out our new file in the JupyterLab CSV viewer!
Now that the data is saved locally, it can be loaded from the local path instead of downloading from the URL:
world2 = pandas.read_csv('data/gapminder_world_data.csv')
world2.head()
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | Asia | Southern Asia | Low | 32.0 | 1040 | 7.57 | 425.0 | 11.9 | NaN | NaN |
| 1 | Afghanistan | 1955 | 8270000 | Asia | Southern Asia | Low | 35.1 | 1130 | 7.52 | 394.0 | 12.7 | NaN | NaN |
| 2 | Afghanistan | 1960 | 9000000 | Asia | Southern Asia | Low | 38.6 | 1210 | 7.45 | 364.0 | 13.8 | NaN | NaN |
| 3 | Afghanistan | 1965 | 9940000 | Asia | Southern Asia | Low | 42.2 | 1190 | 7.45 | 334.0 | 15.2 | NaN | NaN |
| 4 | Afghanistan | 1970 | 11100000 | Asia | Southern Asia | Low | 45.8 | 1180 | 7.45 | 306.0 | 17.0 | 1.36 | 0.21 |
Similar to a dictionary, we can index a specific column of a DataFrame using the column name inside square brackets:
Pro Tip: In Jupyter notebooks, auto-complete works for DataFrame column names!
world['year']
0 1950
1 1955
2 1960
3 1965
4 1970
...
2487 1995
2488 2000
2489 2005
2490 2010
2491 2015
Name: year, Length: 2492, dtype: int64
Note: The numbers on the left are the DataFrame's index, which was automatically added by
pandaswhen the data was loaded
What type of object is this?
type(world['year'])
pandas.core.series.Series
It's a Series, another data structure from the pandas library.
Many of the methods we use on a DataFrame can also be used on a Series, and vice versa
world['year'].head()
0 1950 1 1955 2 1960 3 1965 4 1970 Name: year, dtype: int64
world.year)Select multiple columns:
world[['country', 'year']]
| country | year | |
|---|---|---|
| 0 | Afghanistan | 1950 |
| 1 | Afghanistan | 1955 |
| 2 | Afghanistan | 1960 |
| 3 | Afghanistan | 1965 |
| 4 | Afghanistan | 1970 |
| ... | ... | ... |
| 2487 | Zimbabwe | 1995 |
| 2488 | Zimbabwe | 2000 |
| 2489 | Zimbabwe | 2005 |
| 2490 | Zimbabwe | 2010 |
| 2491 | Zimbabwe | 2015 |
2492 rows × 2 columns
Note the double square brackets!
When you select more than one column, the output is a DataFrame:
type(world[['country', 'year']])
pandas.core.frame.DataFrame
If you'll be frequently using a particular subset, it's often helpful to assign it to a separate variable
populations = world[['country', 'year', 'population']]
populations.head()
| country | year | population | |
|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 |
| 1 | Afghanistan | 1955 | 8270000 |
| 2 | Afghanistan | 1960 | 9000000 |
| 3 | Afghanistan | 1965 | 9940000 |
| 4 | Afghanistan | 1970 | 11100000 |
populations.describe()
| year | population | |
|---|---|---|
| count | 2492.00000 | 2.492000e+03 |
| mean | 1982.50000 | 2.667661e+07 |
| std | 20.15969 | 1.037838e+08 |
| min | 1950.00000 | 2.500000e+04 |
| 25% | 1965.00000 | 1.730000e+06 |
| 50% | 1982.50000 | 5.270000e+06 |
| 75% | 2000.00000 | 1.600000e+07 |
| max | 2015.00000 | 1.400000e+09 |
When selecting a larger number of columns, you may want to define the list of column names as a separate variable
subset_columns = ['country', 'region', 'year', 'population', 'life_expectancy']
world_subset = world[subset_columns]
world_subset.head()
| country | region | year | population | life_expectancy | |
|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 1950 | 7750000 | 32.0 |
| 1 | Afghanistan | Asia | 1955 | 8270000 | 35.1 |
| 2 | Afghanistan | Asia | 1960 | 9000000 | 38.6 |
| 3 | Afghanistan | Asia | 1965 | 9940000 | 42.2 |
| 4 | Afghanistan | Asia | 1970 | 11100000 | 45.8 |
A helpful way to summarize categorical data (such as names of countries and regions) is use the value_counts() method to count the unique values in that column:
world['sub_region'].value_counts()
Sub-Saharan Africa 644 Latin America and the Caribbean 406 Western Asia 252 Southern Europe 168 South-eastern Asia 140 Eastern Europe 140 Northern Europe 140 Southern Asia 126 Western Europe 98 Northern Africa 84 Eastern Asia 70 Central Asia 70 Melanesia 56 Australia and New Zealand 28 Northern America 28 Polynesia 28 Micronesia 14 Name: sub_region, dtype: int64
The output above tells us, for example, that 644 of the observations in our data are for Sub-Saharan Africa (recall that each row is an observation corresponding to a single country in a single year).
value_counts() sorts the output from highest count to lowestsort_index() methodworld['sub_region'].value_counts().sort_index()
Australia and New Zealand 28 Central Asia 70 Eastern Asia 70 Eastern Europe 140 Latin America and the Caribbean 406 Melanesia 56 Micronesia 14 Northern Africa 84 Northern America 28 Northern Europe 140 Polynesia 28 South-eastern Asia 140 Southern Asia 126 Southern Europe 168 Sub-Saharan Africa 644 Western Asia 252 Western Europe 98 Name: sub_region, dtype: int64
If we just want a list of unique values, we can use the unique() method:
world['sub_region'].unique()
array(['Southern Asia', 'Southern Europe', 'Northern Africa',
'Sub-Saharan Africa', 'Latin America and the Caribbean',
'Western Asia', 'Australia and New Zealand', 'Western Europe',
'Eastern Europe', 'South-eastern Asia', 'Northern America',
'Eastern Asia', 'Northern Europe', 'Melanesia', 'Central Asia',
'Micronesia', 'Polynesia'], dtype=object)
If we just want the number of unique values, we can use the nunique() method:
world['sub_region'].nunique()
17
We can extract rows from a DataFrame or Series based on a criteria
For example, suppose we want to select the rows where the life expectancy is greater than 82 years
First we use the comparison operator > on the life_expectancy column:
world['life_expectancy'] > 82
0 False
1 False
2 False
3 False
4 False
...
2487 False
2488 False
2489 False
2490 False
2491 False
Name: life_expectancy, Length: 2492, dtype: bool
The result is a Series with a Boolean value for each row in the data frame indicating whether it is True or False that this row has a value above 82 in the column life_expectancy.
We can find out how many rows match this condition using the sum() method
True is treated as 1 and False as 0.above_82 = world['life_expectancy'] > 82
above_82.sum()
13
We can use a Boolean Series as a filter to extract the rows of world which have life expectancy above 82
df['X'])world[world['life_expectancy'] > 82]
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 111 | Australia | 2015 | 23800000 | Oceania | Australia and New Zealand | High | 82.6 | 43800 | 1.86 | 3.8 | 3.10 | 14.2 | 14.5 |
| 783 | France | 2015 | 64500000 | Europe | Western Europe | High | 82.2 | 37800 | 1.98 | 3.9 | 118.00 | 13.2 | 14.0 |
| 993 | Iceland | 2015 | 330000 | Europe | Northern Europe | High | 82.2 | 42700 | 1.95 | 2.2 | 3.29 | 13.9 | 14.9 |
| 1091 | Italy | 2015 | 59500000 | Europe | Southern Europe | High | 82.3 | 34200 | 1.46 | 3.4 | 202.00 | 13.8 | 14.3 |
| 1118 | Japan | 2010 | 129000000 | Asia | Eastern Asia | High | 82.8 | 35800 | 1.37 | 3.2 | 353.00 | 14.6 | 14.9 |
| 1119 | Japan | 2015 | 128000000 | Asia | Eastern Asia | High | 83.8 | 37800 | 1.44 | 3.0 | 351.00 | 15.1 | 15.5 |
| 1665 | Norway | 2015 | 5200000 | Europe | Northern Europe | High | 82.1 | 63700 | 1.84 | 2.7 | 14.20 | 14.9 | 15.4 |
| 1958 | Singapore | 2010 | 5070000 | Asia | South-eastern Asia | High | 82.7 | 72100 | 1.26 | 2.8 | 7250.00 | 13.3 | 13.0 |
| 1959 | Singapore | 2015 | 5540000 | Asia | South-eastern Asia | High | 83.6 | 80900 | 1.24 | 2.7 | 7910.00 | 14.0 | 13.8 |
| 2071 | Spain | 2015 | 46400000 | Europe | Southern Europe | High | 82.9 | 32200 | 1.35 | 3.4 | 93.00 | 12.8 | 13.5 |
| 2141 | Sweden | 2015 | 9760000 | Europe | Northern Europe | High | 82.1 | 45500 | 1.91 | 2.9 | 23.80 | 14.5 | 15.1 |
| 2154 | Switzerland | 2010 | 7830000 | Europe | Western Europe | High | 82.2 | 55500 | 1.50 | 4.5 | 198.00 | 14.1 | 13.7 |
| 2155 | Switzerland | 2015 | 8320000 | Europe | Western Europe | High | 83.1 | 56500 | 1.54 | 4.1 | 211.00 | 14.6 | 14.4 |
world matching our criteriaWe can use any of the comparison operators (>, >=, <, <=, ==, !=) on a DataFrame column to create Boolean Series for filtering our data
Select data for East Asian countries:
east_asia = world[world['sub_region'] == 'Eastern Asia']
east_asia
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 448 | China | 1950 | 554000000 | Asia | Eastern Asia | Upper middle | 40.7 | 536 | 5.29 | 317.0 | 59.1 | NaN | NaN |
| 449 | China | 1955 | 611000000 | Asia | Eastern Asia | Upper middle | 47.0 | 708 | 5.98 | 291.0 | 65.1 | NaN | NaN |
| 450 | China | 1960 | 658000000 | Asia | Eastern Asia | Upper middle | 30.9 | 891 | 3.99 | 309.0 | 70.1 | NaN | NaN |
| 451 | China | 1965 | 723000000 | Asia | Eastern Asia | Upper middle | 54.3 | 774 | 6.02 | 115.0 | 77.0 | NaN | NaN |
| 452 | China | 1970 | 825000000 | Asia | Eastern Asia | Upper middle | 61.0 | 851 | 5.75 | 114.0 | 87.9 | 5.37 | 3.57 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2039 | South Korea | 1995 | 45300000 | Asia | Eastern Asia | High | 74.2 | 16600 | 1.62 | 10.6 | 466.0 | 12.30 | 11.40 |
| 2040 | South Korea | 2000 | 47400000 | Asia | Eastern Asia | High | 76.0 | 20800 | 1.35 | 7.5 | 487.0 | 13.00 | 12.30 |
| 2041 | South Korea | 2005 | 48700000 | Asia | Eastern Asia | High | 78.3 | 25500 | 1.17 | 5.5 | 501.0 | 13.60 | 13.10 |
| 2042 | South Korea | 2010 | 49600000 | Asia | Eastern Asia | High | 80.1 | 30400 | 1.19 | 4.1 | 510.0 | 14.20 | 13.90 |
| 2043 | South Korea | 2015 | 50600000 | Asia | Eastern Asia | High | 80.9 | 34200 | 1.28 | 3.5 | 520.0 | 14.80 | 14.60 |
70 rows × 13 columns
We can filter the East Asian data further to select the year 2015:
east_asia_2015 = east_asia[east_asia['year'] == 2015]
east_asia_2015
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 461 | China | 2015 | 1400000000 | Asia | Eastern Asia | Upper middle | 76.3 | 13600 | 1.62 | 10.7 | 149.00 | 11.0 | 9.82 |
| 1119 | Japan | 2015 | 128000000 | Asia | Eastern Asia | High | 83.8 | 37800 | 1.44 | 3.0 | 351.00 | 15.1 | 15.50 |
| 1483 | Mongolia | 2015 | 2980000 | Asia | Eastern Asia | Lower middle | 68.2 | 11400 | 2.79 | 18.8 | 1.92 | 10.8 | 12.10 |
| 1651 | North Korea | 2015 | 25200000 | Asia | Eastern Asia | Low | 70.6 | 1390 | 1.92 | 21.1 | 210.00 | 12.5 | 11.60 |
| 2043 | South Korea | 2015 | 50600000 | Asia | Eastern Asia | High | 80.9 | 34200 | 1.28 | 3.5 | 520.00 | 14.8 | 14.60 |
To select rows and columns at the same time, we use the syntax .loc[<rows>, <columns>]:
canada_pop = world.loc[world['country'] == 'Canada', ['country', 'year', 'population']]
canada_pop.head()
| country | year | population | |
|---|---|---|---|
| 392 | Canada | 1950 | 13700000 |
| 393 | Canada | 1955 | 15700000 |
| 394 | Canada | 1960 | 17900000 |
| 395 | Canada | 1965 | 19700000 |
| 396 | Canada | 1970 | 21500000 |
recent_data = world.loc[world['year'] >= 2010,
['country', 'year', 'sub_region', 'population', 'life_expectancy']
]
recent_data.head()
| country | year | sub_region | population | life_expectancy | |
|---|---|---|---|---|---|
| 12 | Afghanistan | 2010 | Southern Asia | 28800000 | 56.2 |
| 13 | Afghanistan | 2015 | Southern Asia | 33700000 | 57.9 |
| 26 | Albania | 2010 | Southern Europe | 2940000 | 76.3 |
| 27 | Albania | 2015 | Southern Europe | 2920000 | 77.6 |
| 40 | Algeria | 2010 | Northern Africa | 36100000 | 76.5 |
&) or any of the criteria (|)and and or to make sure that the comparison occurs for each row in the data frameSelect rows where the sub-region is Northern Europe and the year is 2015:
world.loc[(world['sub_region'] == 'Northern Europe') & (world['year'] == 2015),
['sub_region', 'country', 'year', 'gdp_per_capita']
]
| sub_region | country | year | gdp_per_capita | |
|---|---|---|---|---|
| 615 | Northern Europe | Denmark | 2015 | 45500 |
| 727 | Northern Europe | Estonia | 2015 | 27300 |
| 769 | Northern Europe | Finland | 2015 | 39000 |
| 993 | Northern Europe | Iceland | 2015 | 42700 |
| 1063 | Northern Europe | Ireland | 2015 | 60900 |
| 1231 | Northern Europe | Latvia | 2015 | 23100 |
| 1301 | Northern Europe | Lithuania | 2015 | 27000 |
| 1665 | Northern Europe | Norway | 2015 | 63700 |
| 2141 | Northern Europe | Sweden | 2015 | 45500 |
| 2365 | Northern Europe | United Kingdom | 2015 | 38500 |
Other useful ways of subsetting data include methods such as isin(), between(), isna(), notna()
world.loc[world['country'].isin(['Canada', 'Japan', 'France']) & (world['year'] == 2015),
['country', 'year', 'life_expectancy']
]
| country | year | life_expectancy | |
|---|---|---|---|
| 405 | Canada | 2015 | 81.7 |
| 783 | France | 2015 | 82.2 |
| 1119 | Japan | 2015 | 83.8 |
Data subsets can also be selected using row labels (index), slices (: similar to lists), and position (row and column numbers). For more details, check out this tutorial and the pandas documentation.
a) Create a new DataFrame called americas which contains the rows of world where the region is "Americas" and has the following columns: country, year, sub_region, income_group, pop_density.
b) Use the head() and tail() methods to display the first 20 and last 20 rows.
c) Use the unique() method on the country column to display the list of unique countries in the americas DataFrame.
We can perform calculations with the columns of a DataFrame and assign the results to a new column.
Calculate total GDP by multiplying GDP per capita with population:
world['gdp_total'] = world['gdp_per_capita'] * world['population']
world.head()
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | gdp_total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | Asia | Southern Asia | Low | 32.0 | 1040 | 7.57 | 425.0 | 11.9 | NaN | NaN | 8060000000 |
| 1 | Afghanistan | 1955 | 8270000 | Asia | Southern Asia | Low | 35.1 | 1130 | 7.52 | 394.0 | 12.7 | NaN | NaN | 9345100000 |
| 2 | Afghanistan | 1960 | 9000000 | Asia | Southern Asia | Low | 38.6 | 1210 | 7.45 | 364.0 | 13.8 | NaN | NaN | 10890000000 |
| 3 | Afghanistan | 1965 | 9940000 | Asia | Southern Asia | Low | 42.2 | 1190 | 7.45 | 334.0 | 15.2 | NaN | NaN | 11828600000 |
| 4 | Afghanistan | 1970 | 11100000 | Asia | Southern Asia | Low | 45.8 | 1180 | 7.45 | 306.0 | 17.0 | 1.36 | 0.21 | 13098000000 |
Compute population in millions:
world['pop_millions'] = world['population'] / 1e6
world[['country', 'year', 'population', 'pop_millions']].head()
| country | year | population | pop_millions | |
|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 7750000 | 7.75 |
| 1 | Afghanistan | 1955 | 8270000 | 8.27 |
| 2 | Afghanistan | 1960 | 9000000 | 9.00 |
| 3 | Afghanistan | 1965 | 9940000 | 9.94 |
| 4 | Afghanistan | 1970 | 11100000 | 11.10 |
world.describe()
| year | population | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | gdp_total | pop_millions | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 2492.00000 | 2.492000e+03 | 2492.000000 | 2492.000000 | 2492.000000 | 2492.000000 | 2492.000000 | 1780.000000 | 1780.000000 | 2.492000e+03 | 2492.000000 |
| mean | 1982.50000 | 2.667661e+07 | 62.567135 | 10502.302167 | 4.353752 | 105.670024 | 118.014013 | 7.687713 | 6.959556 | 2.479745e+11 | 26.676613 |
| std | 20.15969 | 1.037838e+08 | 11.518029 | 15478.942158 | 2.049655 | 98.615582 | 372.055683 | 3.242840 | 3.932678 | 9.965413e+11 | 103.783759 |
| min | 1950.00000 | 2.500000e+04 | 23.800000 | 247.000000 | 1.120000 | 2.200000 | 0.502000 | 0.900000 | 0.210000 | 4.000000e+07 | 0.025000 |
| 25% | 1965.00000 | 1.730000e+06 | 54.200000 | 1930.000000 | 2.360000 | 24.000000 | 14.275000 | 5.097500 | 3.600000 | 5.791900e+09 | 1.730000 |
| 50% | 1982.50000 | 5.270000e+06 | 64.650000 | 4925.000000 | 4.340000 | 72.400000 | 44.850000 | 7.635000 | 6.990000 | 2.396160e+10 | 5.270000 |
| 75% | 2000.00000 | 1.600000e+07 | 71.600000 | 12700.000000 | 6.300000 | 167.000000 | 108.000000 | 10.100000 | 10.000000 | 1.261818e+11 | 16.000000 |
| max | 2015.00000 | 1.400000e+09 | 83.800000 | 178000.000000 | 8.870000 | 473.000000 | 7910.000000 | 15.300000 | 15.700000 | 1.904000e+13 | 1400.000000 |
From the summary statistics, we can see that the highest life expectancy in our data is 83.8 years, but we don't know which country and year this is.
We can find out by sorting on the life_expectancy column, from highest to lowest, and displaying the first few rows:
world_sorted_life_exp = world.sort_values('life_expectancy', ascending=False)
world_sorted_life_exp.head()
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | gdp_total | pop_millions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1119 | Japan | 2015 | 128000000 | Asia | Eastern Asia | High | 83.8 | 37800 | 1.44 | 3.0 | 351.0 | 15.1 | 15.5 | 4838400000000 | 128.00 |
| 1959 | Singapore | 2015 | 5540000 | Asia | South-eastern Asia | High | 83.6 | 80900 | 1.24 | 2.7 | 7910.0 | 14.0 | 13.8 | 448186000000 | 5.54 |
| 2155 | Switzerland | 2015 | 8320000 | Europe | Western Europe | High | 83.1 | 56500 | 1.54 | 4.1 | 211.0 | 14.6 | 14.4 | 470080000000 | 8.32 |
| 2071 | Spain | 2015 | 46400000 | Europe | Southern Europe | High | 82.9 | 32200 | 1.35 | 3.4 | 93.0 | 12.8 | 13.5 | 1494080000000 | 46.40 |
| 1118 | Japan | 2010 | 129000000 | Asia | Eastern Asia | High | 82.8 | 35800 | 1.37 | 3.2 | 353.0 | 14.6 | 14.9 | 4618200000000 | 129.00 |
We can see that the highest life expectancy was Japan in 2015, followed closely by Singapore and Switzerland in the same year.
Grouping and aggregation can be used to calculate statistics on groups in the data.
For simplicity, in this section we'll work with the data from year 2015 only.
world_2015 = world[world['year'] == 2015]
world_2015.head()
| country | year | population | region | sub_region | income_group | life_expectancy | gdp_per_capita | children_per_woman | child_mortality | pop_density | years_in_school_men | years_in_school_women | gdp_total | pop_millions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | Afghanistan | 2015 | 33700000 | Asia | Southern Asia | Low | 57.9 | 1750 | 4.80 | 73.2 | 51.7 | 4.13 | 0.98 | 58975000000 | 33.7000 |
| 27 | Albania | 2015 | 2920000 | Europe | Southern Europe | Upper middle | 77.6 | 11000 | 1.71 | 14.0 | 107.0 | 12.00 | 12.30 | 32120000000 | 2.9200 |
| 41 | Algeria | 2015 | 39900000 | Africa | Northern Africa | Upper middle | 77.3 | 13700 | 2.84 | 25.5 | 16.7 | 8.52 | 7.74 | 546630000000 | 39.9000 |
| 55 | Angola | 2015 | 27900000 | Africa | Sub-Saharan Africa | Lower middle | 64.0 | 6230 | 5.77 | 86.5 | 22.3 | 7.24 | 5.31 | 173817000000 | 27.9000 |
| 69 | Antigua and Barbuda | 2015 | 99900 | Americas | Latin America and the Caribbean | High | 77.2 | 20100 | 2.06 | 8.7 | 227.0 | 13.20 | 14.50 | 2007990000 | 0.0999 |
Suppose we want to find the population totals in each region of our data.
Luckily, with pandas there is a better way: using aggregation to compute statistics for groups within our data.
GROUPBY queries in SQL.Aggregation is a "split-apply-combine" technique:
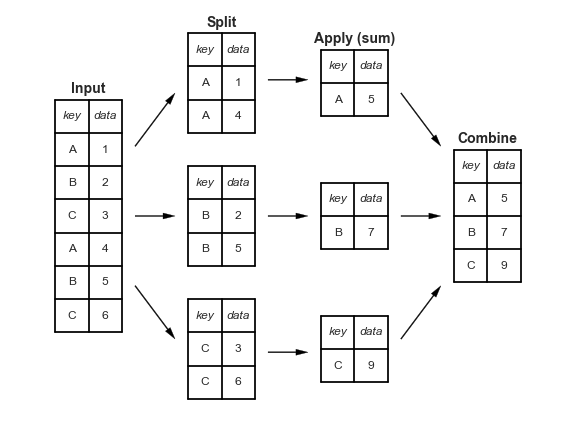
Image credit Jake VanderPlas
For simple aggregations, we can use the groupby() method chained with a summary statistic (e.g., sum(), mean(), median(), max(), etc.)
We will group by region, select the pop_millions column, and take the sum:
world_2015.groupby('region')['pop_millions'].sum()
region Africa 1191.9177 Americas 982.6889 Asia 4391.6350 Europe 740.4830 Oceania 38.4860 Name: pop_millions, dtype: float64
groupby() assigns the variable that we're grouping on (in this case region) to the index of the output dataas_index=False, the grouping variable is instead assigned to a regular columnworld_2015.groupby('region', as_index=False)['pop_millions'].sum()
| region | pop_millions | |
|---|---|---|
| 0 | Africa | 1191.9177 |
| 1 | Americas | 982.6889 |
| 2 | Asia | 4391.6350 |
| 3 | Europe | 740.4830 |
| 4 | Oceania | 38.4860 |
Now let's find the highest population density in each region by aggregating the pop_density column and using max() instead of sum():
world_2015.groupby('region', as_index=False)['pop_density'].max()
| region | pop_density | |
|---|---|---|
| 0 | Africa | 620.0 |
| 1 | Americas | 661.0 |
| 2 | Asia | 7910.0 |
| 3 | Europe | 1340.0 |
| 4 | Oceania | 148.0 |
We can aggregate multiple columns at once. For example, let's find the mean life expectancy and GDP per capita in each region.
world_2015.groupby('region', as_index=False)[['life_expectancy', 'gdp_per_capita']].mean()
| region | life_expectancy | gdp_per_capita | |
|---|---|---|---|
| 0 | Africa | 63.650000 | 5305.961538 |
| 1 | Americas | 75.377419 | 15782.580645 |
| 2 | Asia | 73.772340 | 19755.744681 |
| 3 | Europe | 78.594872 | 31872.307692 |
| 4 | Oceania | 68.900000 | 11932.222222 |
Note: For a more careful analysis, a population-weighted mean would be preferred in the above calculation, to account for the differences in population among countries. Computing a weighted mean within a
pandasaggregation is a bit more involved and beyond the scope of this lesson, so we'll just use the mean.
We can compute sub-totals by grouping on multiple columns:
world_2015.groupby(['region', 'income_group'], as_index=False)['pop_millions'].sum()
| region | income_group | pop_millions | |
|---|---|---|---|
| 0 | Africa | High | 0.0937 |
| 1 | Africa | Low | 543.5870 |
| 2 | Africa | Lower middle | 537.7970 |
| 3 | Africa | Upper middle | 110.4400 |
| 4 | Americas | High | 426.6309 |
| 5 | Americas | Low | 10.7000 |
| 6 | Americas | Lower middle | 32.0500 |
| 7 | Americas | Upper middle | 513.3080 |
| 8 | Asia | High | 246.1000 |
| 9 | Asia | Low | 141.7500 |
| 10 | Asia | Lower middle | 2259.3470 |
| 11 | Asia | Upper middle | 1744.4380 |
| 12 | Europe | High | 493.1250 |
| 13 | Europe | Lower middle | 48.7700 |
| 14 | Europe | Upper middle | 198.5880 |
| 15 | Oceania | High | 28.4100 |
| 16 | Oceania | Lower middle | 8.8840 |
| 17 | Oceania | Upper middle | 1.1920 |
We can use the agg method to compute multiple aggregated statistics on our data, for example minimum and maximum country populations in each region:
world_2015.groupby('region', as_index=False)['pop_millions'].agg(['min', 'max'])
| min | max | |
|---|---|---|
| region | ||
| Africa | 0.0937 | 181.0 |
| Americas | 0.0999 | 320.0 |
| Asia | 0.4180 | 1400.0 |
| Europe | 0.3300 | 144.0 |
| Oceania | 0.1060 | 23.8 |
We can also use agg to compute different statistics for different columns:
agg_dict = {'pop_millions' : 'sum',
'life_expectancy' : ['min', 'max']}
world_2015.groupby('region', as_index=False).agg(agg_dict)
| region | pop_millions | life_expectancy | ||
|---|---|---|---|---|
| sum | min | max | ||
| 0 | Africa | 1191.9177 | 49.6 | 77.4 |
| 1 | Americas | 982.6889 | 63.9 | 81.7 |
| 2 | Asia | 4391.6350 | 57.9 | 83.8 |
| 3 | Europe | 740.4830 | 70.8 | 83.1 |
| 4 | Oceania | 38.4860 | 60.5 | 82.6 |
For even more complex aggregations, there is also a pivot_table() method.
For this exercise we're working with the original DataFrame world (containing all years).
a) Initial setup (you can skip to part b if you've already done this):
world['population'] by 1e6 and assign the result to world['pop_millions']. b) Group the DataFrame world by year and compute the world total population (in millions) in each year.